SimpleTreeLearner is an implementation of classification and regression trees that sacrifices flexibility for speed. A benchmark on 42 different datasets reveals that SimpleTreeLearner is 11 times faster than the original TreeLearner.
The motivation behind developing a new tree induction algorithm from scratch was to speed up the construction of random forests, but you can also use it as a standalone learner. SimpleTreeLearner uses gain ratio for classification and MSE for regression and can handle unknown values.
Comparison with TreeLearner
The graph below shows SimpleTreeLearner construction times on datasets bundled with Orange normalized to TreeLearner. Smaller is better.
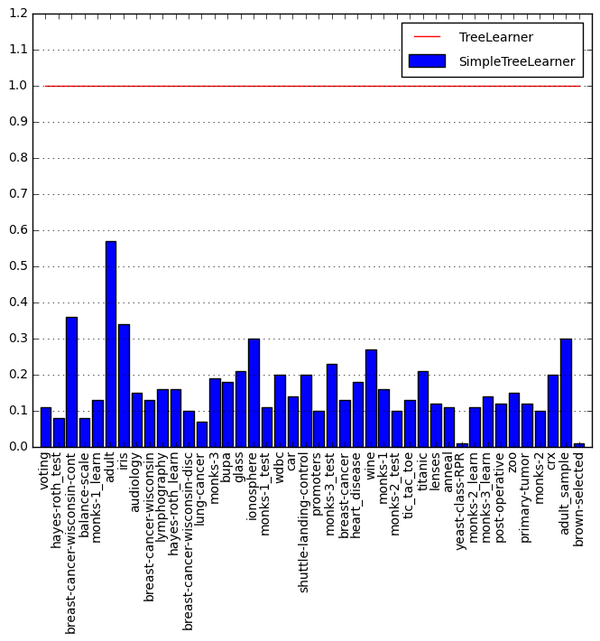
The harmonic mean (average speedup) on all the benchmarks is 11.4.
Usage
The user can set four parameters:
maxMajority
Maximal proportion of majority class.
minExamples
Minimal number of examples in leaves.
maxDepth
Maximal depth of tree.
skipProb
At every split an attribute will be skipped with probability skipProb. This parameter is especially useful for building random forests.
The code snippet below demonstrates the basic usage of SimpleTreeLearner. It behaves much like any other Orange learner would.
import Orange
data = Orange.data.Table("iris")
# build classifier and classify train data
classifier = Orange.classification.tree.SimpleTreeLearner(data, maxMajority=0.8)
for ex in data:
print classifier(ex)
# estimate classification accuracy with cross-validation
learner = Orange.classification.tree.SimpleTreeLearner(minExamples=2)
result = Orange.evaluation.testing.cross_validation([learner], data)
print 'CA:', Orange.evaluation.scoring.CA(result)[0]
